Solution
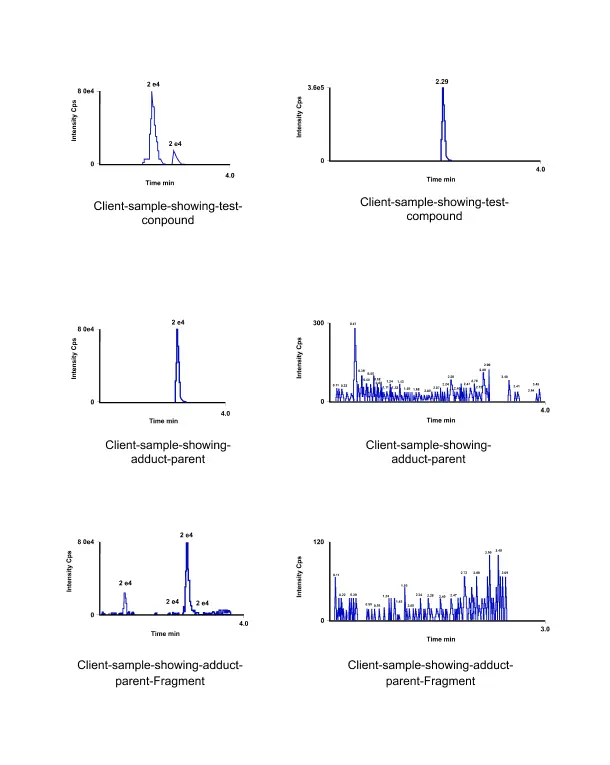
To overcome these challenges, we developed, qualified, and applied an LC-MS/MS method to analyze DNA adduct analysis in cell-based in-vitro samples. The method involved the following steps: (1) isolation of DNA from cultured cells using a commercial kit, which ensured high yield and purity of DNA; (2) enzymatic hydrolysis of DNA to nucleosides, which reduced the complexity of the sample and increased the sensitivity of the analysis; (3) online solid-phase extraction and separation of nucleosides by reversed-phase chromatography, which removed the interference of cell culture components and achieved good resolution of the analytes; (4) detection and identification of DNA adducts by a high-resolution mass spectrometer, which was operated in positive electrospray ionization mode and data-independent acquisition mode, which enabled the simultaneous acquisition of precursor and fragment ion spectra for all the analytes. The method was qualified according to the fit-for-purpose approach, which assessed the method’s performance based on the intended use and the acceptance criteria. The method demonstrated acceptable performance for detecting and quantifying DNA adducts in cell-based in-vitro samples in terms of accuracy, precision, linearity, selectivity, and stability. The method was applied for DNA adducts analysis induced by different genotoxic compounds, such as benzo[a]pyrene, aflatoxin B1, and acetaldehyde, in various cell lines, such as HepG2, A549 and TK6, which represented different organs and tissues.